Sidebar
Approvals: 0/1
sp_rviper
Initial 3D Model - RVIPER: Reproducible ab initio 3D structure determination. The program determines a validated initial intermediate resolution structure using a subset of class averages produced by ISAC2.
Usage
Usage in command line
sp_rviper.py stack directory --radius=outer_radius --sym=sym --n_rv_runs=n_rv_runs --iteration_start=iteration_start --n_v_runs=n_v_runs --npad=npad --criterion_name=criterion_name --outlier_index_threshold_method=outlier_index_threshold_method --angle_threshold=angle_threshold --outlier_percentile=outlier_percentile --ir=inner_radius --rs=ring_step --xr=x_range --yr=y_range --ts=translational_search_step --delta=angular_step --center=center_type --maxit1=max_iter1 --maxit2=max_iter2 --mask3D=mask3D --moon_elimination=moon_elimination --L2threshold=L2threshold --ref_a=ref_a --n_shc_runs=n_shc_runs --doga=doga --fl=fl --aa=aa --pwreference=pwreference --theta1=theta1 --theta2=theta2 --dpi=dpi
Typical usage
sp_rviper exists only in MPI version.
mpirun --npernode 16 -np 48 --host node1,node2,node3 sp_rviper.py stack output_directory --radius=outer_radius --outlier_percentile=95 --fl=0.25 --xr=2 --moon_elimination=750,4.84
The RVIPER exists only in MPI version. Importantly, the number of used MPI processes must be a multiple of --n_shc_runs (default = 4).
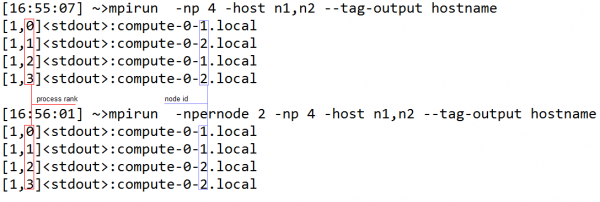
Since RVIPER uses group of processors working together, it is important for efficient execution to have processors within a group allocated to the same node. This way any data exchange within the group does not involve network traffic. The --npernode option of mpirun accomplishes this goal. As shown in the example below when --npernode is used MPI allocates the ranks of the processors sequentially, not moving to the next node until the current one is filled. If --npernode is not used then processors are allocated in a round robin fashion (i.e. jumping to the next node with each allocation). Since in VIPER, groups contain consecutively ranked processors, it is important to provide “--npernode XX”, where XX is the number of processors per node.
Input
Main Parameters
- stack
- Input images stack: A small subset of class averages produced by ISAC2. (default required string)
- directory
- Output directory: The automatically-created output directory will contain results. If the directory already exists, results will be written there, possibly overwriting previous runs. (default required string)
- --radius
- Particle radius [Pixels]: Use the same value as in ISAC2. It has to be less than half the box size. (default 29)
- --sym
- Point-group symmetry: Point-group symmetry of the particle. (default c1)
Advanced Parameters
- --moon_elimination
- Eliminate disconnected regions: Used to removed disconnected pieces from the model. As an argument it requires a comma-separated string with the mass in KDa and the pixel size in [A]. (default none)
- --iteration_start
- Restarting iteration: Iteration from which to restart the program. 0 means go to the most recent one. (default 0)
- --n_rv_runs
- RVIPER iterations: Corresponds to main### output directory. (default 10)
- --n_v_runs
- Minimun number of VIPER runs per RVIPER iterations: Corresponds to run### output directory. (default 3)
- --criterion_name
- Stable projection criterion: Used to decide if the solution is stable, i.e., whether projection images adopt similar orientations in independent runs of the program. Valid options are: '80th percentile', or 'fastest increase in the last quartile'. (default 80th percentile)
- --outlier_index_threshold_method
- Outlier selection method: Used to decide which images to keep. Valid options are: 'discontinuity_in_derivative', 'percentile', or 'angle_measure'. (default discontinuity_in_derivative)
- --angle_threshold
- Angle threshold: Threshold used to remove projections if 'angle_measure' is used to decide the outliers. (default 30)
- --outlier_percentile
- Percentile for outlier: Threshold above which images are considered outliers and removed if 'percentile' is used as outlier selection method. (default 95.0)
- --ir
- Inner rotational search radius [Pixels]: Inner rotational search radius [Pixels]. (default 1)
- --rs
- Ring step size [Pixels]: Step between rings used for the rotational search. (default 1)
- --xr
- X search range [Pixels]: The translational search range in the x direction. Search will +/-xr range in steps of ts. (default '0')
- --yr
- Y search range [Pixels]: The translational search range in the y direction. If omitted it will be xr. (default '0')
- --ts
- Translational search step [Pixels]: The search will be performed in -xr, -xr+ts, 0, xr-ts, xr, can be fractional. (default '1.0')
- --delta
- Projection angular step [Degrees]: Projection angular step. (default '2.0')
- --center
- Center 3D template: -1: center of coordinates, 0: no centering; 1: center of gravity (default -1.0)
- --maxit1
- Maximum iterations - GA step: Maximum number of iterations for GA step. (default 400)
- --maxit2
- Maximum iterations - Finish step: Maximum iterations number of for Finish step. (default 50)
- --mask3D
- 3D mask: Path to 3D mask file. (default sphere)
- --L2threshold
- GA stop threshold: Defines the maximum relative dispersion of structures' L2 norms. (default 0.03)
- --ref_a
- Projection generation method: Method for generating the quasi-uniformly distributed projection directions. S - Saff algorithm, or P - Penczek 1994 algorithm. (default S)
- --n_shc_runs
- GA population size: This defines the number of quasi-independent structures generated. (same as --nruns parameter from sp_viper). (default 4)
- --doga
- Threshold to start GA: Do GA when the fraction of orientation that changes less than 1.0 degrees is at least this fraction. (default 0.1)
- --fl
- Low-pass filter frequency [1/Pixels]: Using a hyperbolic tangent low-pass filter. Specify with absolute frequency. (default 0.25)
- --aa
- Low-pass filter fall-off [1/Pixels]: Fall-off of for the hyperbolic tangent low-pass filter. Specify with absolute frequency. (default 0.1)
- --pwreference
- Power spectrum reference: Text file containing a 1D reference power spectrum. (default none)
- --npad
- Image padding factor: Used by 3D reconstruction algorithm in the program. (default 2)
- --theta1
- Minimum theta: Lower bound for out-of-plane tilt angle. No tilt corresponds to 90 degrees. (default -1)
- --theta2
- Maximum theta: Upper bound for out-of-plane tilt angle. No tilt corresponds to 90 degrees. (default -1)
- --dpi
- BILD resolution: Resolution in dpi for angular distribution plot. (default 72)
Output
The output directory structure generated by sp_rviper is shown in the figure below. Each runXXX directory contains the output of running the VIPER algorithm (please see sp_viper). The runXXX directory contains the reconstructed structure of stage1, refvolf2.hdf, and parameters into refparams2.txt. After stage 2, the final structure and parameters will be written to volf.hdf and params.txt. Other output files are log.txt and previousmax.txt. Each mainXXX directory contains the output of --n_v_runs viper runs (default 3). The number of mainXXX directories is given by --n_rv_runs.
Description
Method
This program uses multiple VIPER runs to find unstable projections. Based on the user chosen criterion it eliminates the unstable projections and reruns again until all projections are stable. Since VIPER is used as a building block, all requirements from VIPER must be satisfied. Attributes xform.projection have to be set in the header of each file. If their values are not known, all should be set to zero. Determining whether the --n_v_runs reconstructed maps in the current RVIPER iteration have a core set of stable projections is done using one of the following criteria shown in the figures below. The y axis represents the error angle. For example, if a projection has the following assigned angles in three different reconstructed maps 30,45 and 55 then the error associated with this image is abs(30-45) + abs(30-55) + abs(45-55))/3 = 16.6. The x axis represents the image index of the sorted array of error angles.
The first criterion, called “80th percentile” (left image) is satisfied when the 80th percentile is less or equal to 20% of the maximum. The second criterion, called “fastest increase in the last quartile” is satisfied when the last quartile has a length greater than 20% of the maximum. If finishing criterion is not met after executing 10 VIPER runs, (the criterion fails for all combinations of --n_v_runs (default=3) taken by 10 (120 in total)) then the program stops.
Once a criterion is met, a decision is made regarding which images to keep. Currently there are three options implemented:
- Percentile: All images (sorted by their angle error) with error below “outlier_percentile” given in the command line are kept for the next iteration.
- Angle_measure: All images that have angle error below “angle_threshold” given in the command line are kept for the next iteration.
- Discontinuity_in_derivative: As shown in the figure below, two lines (green and red) are fitted together against the error curve (blue) while their common point moves along the x axis between 80th percentile and “outlier_percentile” (provided in the command line). The point on the x coordinate where the projections of the best fit lines meet is chosen as the outlier index threshold. All images before it are kept for the next iteration.
Time and Memory
On our cluster, it takes about 6 hours to process 400 88×88 particles on 64 processors. Memory required is about 0.5GB per processor.
Example of RVIPER output
In the example below, RVIPER found in the third iteration (main003) a set of 3 structures with stable angle assignments. Based on the three structures the program generates variance_volume.hdf and average_volume.hdf which can be used as an initial reference.
Reference
Penczek 1994, “The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles”, Ultramicroscopy 53, 251-270.
Developer Notes
Author / Maintainer
Horatiu Voicu, Pawel A. Penczek
Keywords
Category 1:: APPLICATIONS Category 3:: GRIDDING
Files
sparx/bin/sp_rviper.py
See also
Maturity
Beta:: Under evaluation and testing. Please let us know if there are any bugs.
Bugs
There are no known bugs so far.