Sidebar

Table of Contents
Cinderella: Deep learning based binary classification tool
From the fairy tale Cinderella
Our binary classification tool (Cinderella) is based on a deep learning network to classify class averages, micrographs or subtomograms into good and bad categories.
Cinderella supports .hdf/.mrcs files for class averages, .mrc files for micrographs, and .hdf files for subtomograms.
Cinderella was written to automate cryo-em data processing. It's open source and easy to use.
We provide a pretrained general model for classifying class averages (see tutorial). But you can easily train it with your own set of classes, micrographs, and/or subtomograms.
2D class selection model
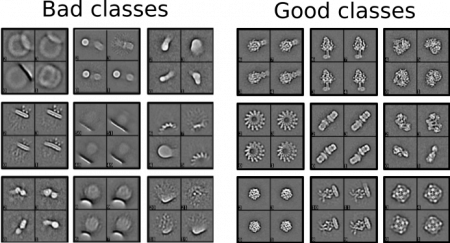
Our model was trained on a set of 2D classes from both ISAC and Relion. During the creation of the training data set, we tried to answer the question, “Which class would I select If I would not know the particle?” when deciding what is a “good” class. Here are a couple of examples for good/bad classes in Cinderella:
You can easily contribute your own classes!
Right now our model is trained on 4773 good classes and 5390 bad classes.
Download
Cinderella
Pretrained model (2D classes)
Uploaded: 27. August 2020, Dataset: 4773 good classes and 5390 bad classes.
Archive
Old versions of cinderella and the pretrained model can be found in the archive.
Changelog
Version 0.7
- Now uses a circular masks by default. This allows to use full rotation during data augmentation. Can be deactivated by setting the field
mask_radiusin the configuration file to -1. In case you want to use an model trained with Cinderella < 0.7 please set the radius to -1. Otherwise you case specify any radius you want. By default (nomask_radiusprovided) it will use 0.4*input_size. - The general models now includes 300 new good Relion classes and 2000 new bad Relion classes (Thanks to Takanori Nakane and Grigory Sharov).
- Fixed numerical instability that occurs when you have classes filled with a constant value (Thanks to Grigory Sharov).
- Fixed a problem with classes that contain NaN values. NaN values are now replaced with 0. (Thanks to Grigory Sharov).
- Fixed an issue when filenames contain more than one point.
Version 0.6
- Fix an issue for classes in mrcs format
- Minor changes
Version 0.5
- Add support for subtomograms
- Faster file reading
Version 0.4
- Balances unbalanced training datasets.
- It is now possible to train Cinderella to select micrographs
- Updated the general model for 2D class selection.
Version 0.3.1
- Downgrade to tensorflow 1.10.1 again, as user report long initialization times
- Only report the number of good / bad classes + their fraction.
Version 0.3.0
- More data augmentation (add rotation)
- Better sampling of validation data. It is now ensured that each file contributes some validation data.
- Updated tensorflow to 1.12.3 to make it compatible to the crYOLO environment
Contribute
Here is the repository of our training data:
Download the public training data
Unfortunately, we cannot upload the complete training dataset, as some classes are from projects that are not yet published.
If you want to contribute with your own classes, please upload them here:
Ideally, please upload separate HDF/mrcs files for good and bad classes. You can do this separation with EMAN2's e2display. However, you can also upload the classes without separation and we will try to do it.
Installation
The following instructions assume that pip and anaconda or miniconda are available. In case you have a old cinderella environment installed, first remove the old one with:
conda env remove --name cinderella
After that, create a new virtual environment:
conda create -n cinderella -c anaconda python=3.6 pyqt=5 cudnn=7.1.2 numpy==1.14.5
Activate the environment:
source activate cinderella
Install Cinderella for GPU:
pip install cinderella[gpu]
… or CPU:
pip install cinderella[cpu]