Sidebar
 This version is outdated by a newer approved version.
This version is outdated by a newer approved version. This version (2020/09/23 13:59) was approved by fschoenfeld.The Previously approved version (2020/09/23 13:39) is available.
This version (2020/09/23 13:59) was approved by fschoenfeld.The Previously approved version (2020/09/23 13:39) is available.
This is an old revision of the document!
Table of Contents
Overview
ISAC (Iterative Stable Alignment and Clustering) is a 2D classification algorithm. It sorts a given stack of cryo-EM particles into different classes that share the same view of a target protein. ISAC is based around iterations of alternating equal size k-means clustering and repeated 2D alignment routines.
Yang, Z., Fang, J., Chittuluru, J., Asturias, F. J. and Penczek, P. A. (2012) Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure 20, 237–247.
ISAC versions
- ISAC is the initial version as described in the original paper. At this point this implementation is obsolete and has been replaced by ISAC2 and GPU ISAC (see below).
- ISAC2 is an improved version of ISAC and used by default to produce 2D class averages in the SPHIRE (git) software package and the TranSPHIRE automated pipeline for processing cryo-EM data. ISAC2 is a CPU-only implementation and was developed to run on a computer cluster.
- GPU ISAC was developed to run ISAC2 on a single workstation by outsourcing its computationally expensive bottleneck calculations to any available GPUs, while simultaneously keeping its MPI-based CPU parallelization otherwise intact. GPU ISAC is provided as an add-on to SPHIRE that can be installed manually (see below).
Download & Installation
- CUDA: These installation instructions assume that CUDA is already installed on your system. You can confirm this by running
nvcc --versionin your terminal; the resulting output should list the version of your installed CUDA compilation tools.
- SPHIRE: In order to use GPU ISAC, SPHIRE needs to be installed. You can find the SPHIRE download and installation instructions here. You can confirm a working SPHIRE version by running
which sphirein your terminal; the resulting output should give you the path to your SPHIRE installation (the path should indicate a version number of 1.3 or higher).
Download
- GPU ISAC is currently developed as a manually installed add-on for SPHIRE and distributed as a .zip file that can be found here: GPU ISAC (v2.3.2) download link.
Installation
Before you start, make sure your SPHIRE environment is activated.
How to activate your SPHIRE environment:
How to activate your SPHIRE environment:
- During the SPHIRE installation, an Anaconda environment for SPHIRE was created. You can list your available Anaconda environments using:
conda env list
- Look for your SPHIRE environment and activate it using either:
conda activate NAME_OF_YOUR_ENVIRONMENT
or
source activate NAME_OF_YOUR_ENVIRONMENT
It will depend on your system and Anaconda installation which one of these you will have to use.
GPU ISAC comes with a handy installation script that can be used as follows:
- Extract the archive to your chosen GPU ISAC installation folder.
- Open a terminal and navigate to your installation folder.
- Run the installation script:
./install.sh
All done!
Running GPU ISAC
An example call to use GPU ISAC looks as follows:
mpirun python /path/to/sxisac2_gpu.py bdb:path/to/stack path/to/output --CTF -–radius=160 --img_per_grp=100 --minimum_grp_size=60 --gpu_devices=0,1
Using the following mix of both mandatory and optional parameters (see below to learn which is which):
mpirun python /path/to/sxisac2_gpu.py bdb:path/to/stack path/to/output --CTF -–radius=160 --img_per_grp=100 --minimum_grp_size=60 --gpu_devices=0,1
[ ! ] - Mandatory parameters in the GPU ISAC call:
mpirunis not a GPU ISAC parameter, but is required to launch GPU ISAC using MPI parallelization (GPU ISAC uses MPI to parallelize CPU computations and MPI/CUDA to distribute and parallelize GPU computations)./path/to/sxisac2_gpu.pyis the path to your sxisac2_gpu.py file. If you followed these instructions it should beyour/installation/path/gpu_isac_2.2/bin/sxisac2_gpu.py.path/to/stackis the path to your input .bdb stack. If you prefer to use an .hdf stack, simply remove thebdb:prefix.path/to/outputis the path to your preferred output directory.--radius=160is the radius of your target particle (in pixels) and has to be set accordingly.--gpu_devicestells GPU ISAC what GPUs to use by specifying their system id values.
What GPUs do I have and what are their system id values?
What GPUs do I have and what are their system id values?
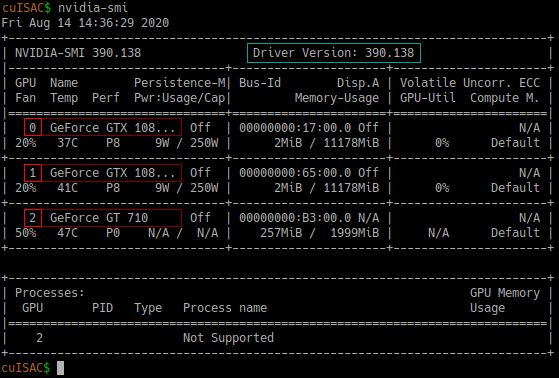
You can use nvidia-smi in your terminal to see what GPUs are available on your machine. This also lists their id values and sorts all entries by CUDA compute capability, where your most powerful GPU has id value 0 and your least powerful GPU has the highest id value:
Above: Example output of nvidia-smi. GPU system id values and GPU names marked in red. Among other things, this also lists your current driver version, marked in turquoise.
[?] - Optional parameters recommended to be used when running GPU ISAC:
- Use the
--CTFflag to apply phase flipping to your particles. - Use
--img_per_grpto limit the maximum size of individual classes. Empirically, a class size of 100-200 (30-50 for negative stain) particles has been proven successful when dealing with around 100,000 particles. (This may differ for your data set and you can use GPU ISAC to find out; see below.) - Use
--minimum_grp_sizeto limit the minimum size of individual classes. In general, this value should be around 50-60% of your maximum class size.
- An up to date list of all GPU ISAC parameters can always be printed by using the
-hparameter (in this case you do not need to specify any other parameters):mpirun python /path/to/sxisac2_gpu.py -h
or simply
python /path/to/sxisac2_gpu.py -h
- The online documentation of ISAC2 parameters can be found here.
- Additional utilities that are helpful when using any version of ISAC can be found here.
- More information about using ISAC for 2D classification can also be found in the ISAC chapter of the official SPHIRE tutorial (link to .pdf file).
Examples
EXAMPLE 01: Test run
This example is a test run that can be used to confirm GPU ISAC was installed successfully. It is a small stack that contains 64 artificial faces and is already included in the GPU ISAC installation package. You can process it using GPU ISAC as follows:
- In your terminal, navigate to your GPU ISAC installation folder:
cd /gpu/isac/installation/folder
- Run GPU ISAC:
mpirun python bin/sxisac2_gpu.py 'bdb:examples/isac_dummy_data_64#faces' 'isac_out_test/' --radius=32 --img_per_grp=8 --minimum_grp_size=4 --gpu_devices=0
Note that we don't care about the quality of any produced averages here; this test is used to make sure there are no runtime issues before a more time consuming run is executed.
EXAMPLE 02: TcdA1 toxin data
This example uses the SPHIRE tutorial data set (link to .tar file) described in the SPHIRE tutorial (link to .pdf file). The data contains about 10,000 particles from 112 micrographs and was originally published here (Gatsogiannis et al, 2013).
After downloading the data you'll notice that the extracted folder contains a multitude of subfolders. For the purposes of this example we are only interested in the Particles/ folder that stores the original data as a .bdb file.
You can process this stack using GPU ISAC as follows:
- In your terminal, navigate to your GPU ISAC installation folder:
cd /gpu/isac/installation/folder
- Run GPU ISAC:
mpirun python bin/sxisac2_gpu.py 'bdb:/your/path/to/Particles/#stack' 'isac_out_TcdA1' --CTF --radius=145 --img_per_grp=100 --minimum_grp_size=60 --gpu_devices=0
- Replace
/your/path/to/Particles/with the path to theParticles/directory you just downloaded. - Optional: Replace
--gpu_devices=0with--gpu_devices=0,1if you have two GPUs available (and so on).
The final averages can then be found in isac_out_TcdA1/ordered_class_averages.hdf. You can look at them using e2display.py (or any other displaying program of your choice) and should see averages like these:
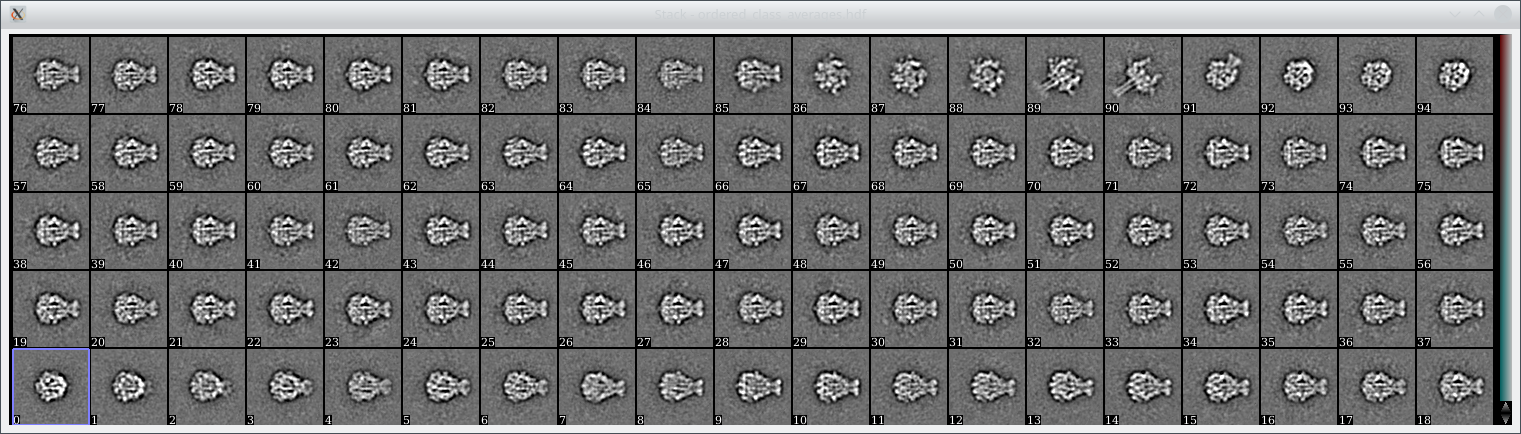 Above: 95 class averages produced when processing the above data set using GPU ISAC. The particle stack contains 11,003 particles and the averages were computed within 6 minutes (Intel i9-7020X CPU and 2x GeForce GTX 1080 GPUs).
Above: 95 class averages produced when processing the above data set using GPU ISAC. The particle stack contains 11,003 particles and the averages were computed within 6 minutes (Intel i9-7020X CPU and 2x GeForce GTX 1080 GPUs).
Usage
Next to producing high quality 2D class averages, GPU ISAC is also an excellent tool to screen your data which allows you to:
- Quickly generate 2D class averages.
- Quickly identify suitable parameters for your data set.
- Quickly gauge the quality of your data set before spending time on more costly processing steps.
Well, “suitable parameters” sounds great! How do I get those?
Well, “suitable parameters” sounds great! How do I get those?
Clustering cryo-EM data is a difficult problem that involves many different parameters and often it is unclear how these impact the resulting 2D class averages. In GPU ISAC the most relevant parameters to fiddle with are:
- Class size: The class (or cluster) size
--img_per_grpin ISAC determines how many particles are taken together in order to construct a new 2D class average. High values will mean cleaner averages, but might also lump together particles that should be sorted into different classes. If you are using GPU ISAC to screen a set of 20,000 to 40,000 particles, then '100' particles per class are a good starting value. Further, the minimun size of each class--minimum_grp_sizeshould be around 60% of set class size. - Threshold error: The
--thld_errparameter determines how similar subsequently produced averages have to be in order to be considered stable enough. A value of0.7is very stringent, while1.4is less so, and you should not need a higher value than2.4.
Since GPU ISAC processes small stacks of about 10,000 to 20,000 particles fairly quickly, you can try several runs with different values for --img_per_group and --thld_err to see which combination gives you the best results.
Once you are happy with the results, you can use these parameters for a full-sized run of (GPU) ISAC. Good luck! :)
GPU ISAC output files
GPU ISAC produces a multitude of output files that can be used to analyze the success of running the program, even while it is still ongoing. These include the following:
- Main iteration folders: As GPU ISAC is running, it performs multiple “main iterations” and “generations” that are stored within the output folder structure. New class averages are produced during every iteration / generation and can be looked at during runtime without having to wait for the overall process to conclude. This can help to quickly gauge the quality of a data set. Check
path/to/output/mainXXX/generationYYYfor the.hdffiles to that contain any newly produced class averages. - In both the main iteration folders and the base output folder you will find
processed_images.txtfiles. These contain the indices of all processed particles and can be used to determine how many particles GPU ISAC did account for during classification. - The final averages are stored in
path/to/output/ordered_class_averages.hdf.
Release notes
GPU ISAC limitations
- The current develpoment goal of GPU ISAC is to run as fast as possible on a single machine. Because of this priority, GPU ISAC does not yet run on multiple nodes. This is planned to change as soon as the currently known bottlenecks have all been converted to run on the available GPUs.
Known issues
- In some cases when using CUDA version 11, GPU ISAC receives a kill signal interrupt. We're investigating the issue but recommend to use a lower version (confirmed working when using CUDA 9 and 10) until it is resolved. You can use
nvcc –versionin your terminal to see the CUDA version you are using.
GPU ISAC v2.3.1 & v2.3.2 (hotfix releases)
- Changed input file handling, which results in a massive reduction in memory use.
- Fixed use of
-hparameter to display the help. - Fixed error in the pre-alignment progress bar that made it seem as if it did not run to completion.
- Minimum class size is now 60% of the full class size, if no minimum class size was specified by the user.
GPU ISAC v2.3
- Updated GPU ISAC install package for version 2.3.
- Fixed multiple issues occurring when handling larger data sets (200k and upwards).
- Added memory checks to the output to document GPU ISAC memory use.
- Batched input read, part I: Input is now read in batches with pre-processing already applied during reading. This means that we can now deal with data sets that do not fit into system RAM (as long as the compressed data does).
- Batched input read, part II: Input reading and processing is spread across all MPI processes, while actual processing (pre-alignment) is spread only across GPU processes. This ensures data is processed using all CPU & GPU resources available to the used machine.
- Multiple bug fixes for increased stability and functionality.
- Cleanup of old code for increased readability and maintainability.
GPU ISAC v2.2
- Created an easy installation package for GPU ISAC including a quick test to confirm a successful installation.
- Added more sanity checks to catch invalid parameter combinations and abort the run immediately.
- Updated the transformation stack in GPU alignment functions for higher quality averages.
- Multiple bug fixes for increased stability and functionality.
GPU ISAC “Chimera”
- Initially released beta version of GPU ISAC.