Sidebar
 This version (2019/09/18 17:08) was approved by twagner.The Previously approved version (2019/09/18 10:19) is available.
This version (2019/09/18 17:08) was approved by twagner.The Previously approved version (2019/09/18 10:19) is available.
This is an old revision of the document!
Table of Contents
Picking filaments - Using a model trained for your data
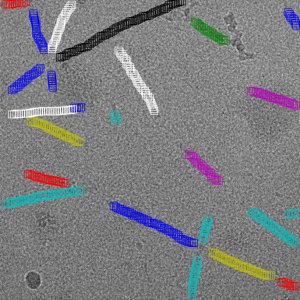
When picking filaments, it is important to identify each filament individually. This allows specific spacing of the boxes (i.e., the helical rise) to maximize the number of particles. CrYOLO supports this method of picking filaments.
Filament mode on actin:
Filament mode on MAVS (EMPIAR-10031) :
If you followed the installation instructions, you now have to activate the crYOLO virtual environment with
source activate cryolo
1. Data preparation
The first step is to create the training data for your model. Right now, you have to use the e2helixboxer.py for this:
e2helixboxer.py --gui train_image/*.mrc
After tracing your training data in e2helixboxer, export them using File → Save. Unfortunately you have to do that with each image separately.
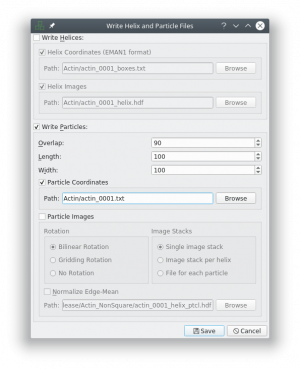
Make sure that you uncheck the boxes “Write Helices” and “Particle Images” and check the box “Particle Coordinates”, as this the only format supported right now (see screenshot). Also remove the “_helix_ptcl_coords” suffix in the path field. The coordinate files have to have the same name as the micrographs.
In the following example, it is expected that you exported into a folder called “train_annot”.
For projects with roughly 20 filaments per image we successfully trained on 40 images (⇒ 800 filaments).
2. Start crYOLO
You can use crYOLO either by command line or by using the GUI. The GUI should be easier for most users. You can start it with:
cryolo_gui.py
The crYOLO GUI is essentially a visualization of the command line interface. On left side, you find all possible “Actions”:
- config: With this action you create the configuration file that you need to run crYOLO.
- train: This action lets you train crYOLO from scratch or refine an existing model.
- predict: If you have a (pre)trained model you can pick particles in your data set using this command.
- evaluation: This action helps you to quantify the quality of your model.
- boxmanager: This action starts the cryolo boxmanager. You can visulize the picked particles with it or create training data.
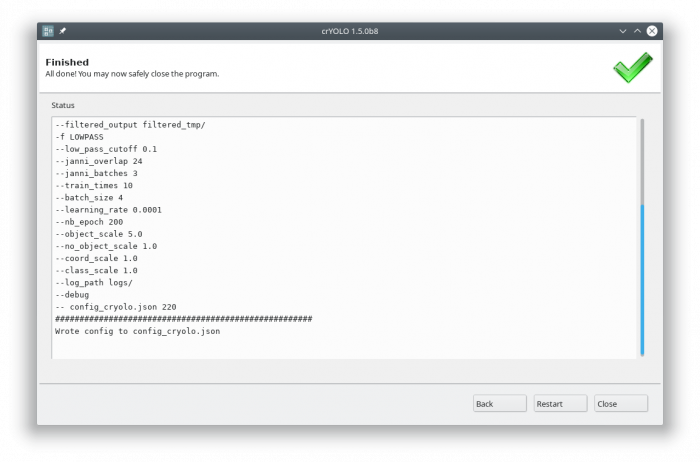
Each action has several parameters which are organized in tabs. Once you have chosen your settings you can press [Start] (just as example, don't press it now ![]() ), the command will be applied and crYOLO shows you the output:
), the command will be applied and crYOLO shows you the output:
It will tell you if something went wrong. Moreover, it will tell you all parameters used. Pressing [Back] brings you back to your settings, where you can either edit the settings (in case something went wrong) or go to the next action.
3. Configuration
You now have to create a configuration file for your picking project. It contains all important constants and paths and helps you to reproduce your results later on.
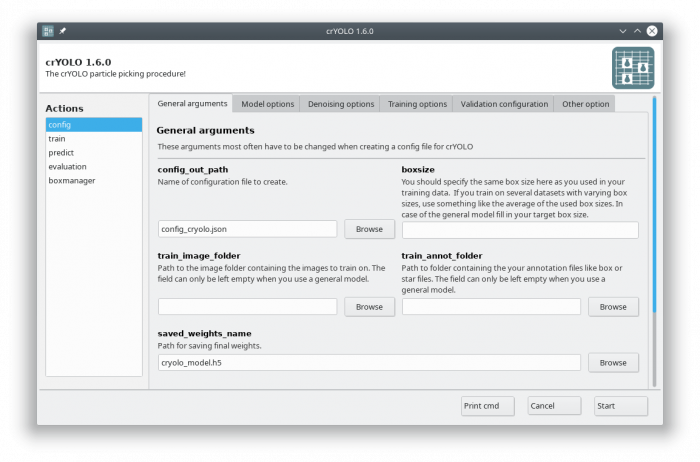
You can either use the command line to create the configuration file or the GUI. For most users, the GUI should be easier. Select the config action and fill in the general fields:
At this point you could already press the [Start] button to generate the config file but you might want to take these options into account:
- During training, crYOLO also needs validation data1). Typically, 20% of the training data are randomly chosen as validation data. If you want to use specific images as validation data, you can move the images and the corresponding box files to separate folders. Make sure that they are removed from the original training folder! You can then specify the new validation folders in “Validation configuration” tab.
- By default, your micrographs are low pass filtered to an absolute frequency of 0.1 and saved to disk. You can change the cutoff threshold and the directory for filtered micrographs in the “Denoising options” tab.
- When training from scratch, crYOLO is initialized with weights learned on the ImageNet training data (transfer learning2)). However, it might improve the training if you set the pretrained_weights options in the “Training options” tab to the current general model. Please note, doing this you don't fine tune the network, you just change the initial model initialization.
Since crYOLO 1.4 you can also use neural network denoising with JANNI. The easiest way is to use the JANNI's general model (Download here) but you can also train JANNI for your data. crYOLO directly uses an interface to JANNI to filter your data, you just have to change the filter argument in the Denoising tab from LOWPASS to JANNI and specify the path to your JANNI model:
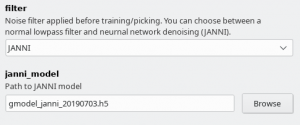 I recommend to use denoising with JANNI only together with a GPU as it is rather slow (~ 1-2 seconds per micrograph on the GPU and 10 seconds per micrograph on the CPU)
I recommend to use denoising with JANNI only together with a GPU as it is rather slow (~ 1-2 seconds per micrograph on the GPU and 10 seconds per micrograph on the CPU)
You can also modify all options and parameters directly in the config.json file. It can be opened by any text editor. Please note the wiki entry about the crYOLO configuration file if you want to know more details.
Click to display ⇲
Click to hide ⇱
To create a basic configuration file that will work for most projects is very simple. I assume your box files for training are in the folder train_annot and the corresponding images in train_image. I furthermore assume that your box size in your box files is 160. To create the config config_cryolo.json simply run:
cryolo_gui.py config config_cryolo.json 160 --train_image_folder train_image --train_annot_folder train_annot
To get a full description of all available options type:
cryolo_gui.py config -h
If you want to specify separate validation folders you can use the --valid_image_folder and --valid_annot_folder options:
cryolo_gui.py config config_cryolo.json 160 --train_image_folder train_image --train_annot_folder train_annot --valid_image_folder valid_img --valid_annot_folder valid_annot
4. Training
Now you are ready to train the model. In case you have multiple GPUs, you should first select a free GPU. The following command will show the status of all GPUs:
nvidia-smi
For this tutorial, we assume that you have either a single GPU or want to use GPU 0.
In the “Optional arguments” tab you can change the GPU that should be used by crYOLO. If you have multiple GPUs (e.g. nvidia-smi lists GPU 0 and GPU 1) you can also use both by setting the GPU argument to '0 1'.
In the GUI you have to fill in the mandatory fields:
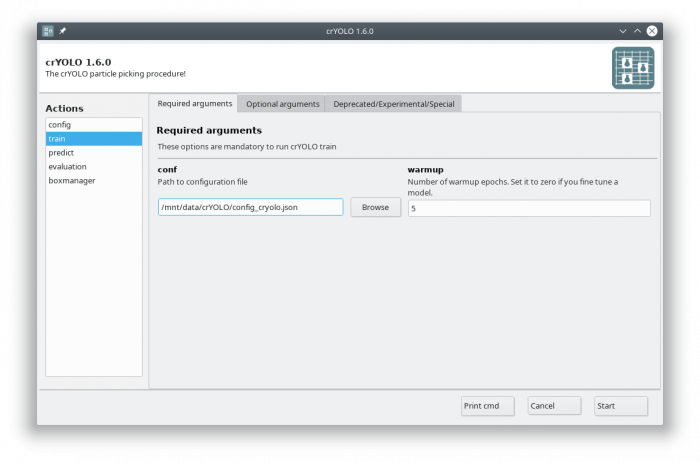
The default number of warmup epochs3) is fine as long as you don't want to refine an existing model. During the warmup training epochs it will not try to estimate the size of your particle, which helps crYOLO to converge.
When you start the training, it will stop when the “loss” metric on the validation data does not improve 10 times in a row. This is typically enough. In case you want to give the training more time to find the best model can increase the “not changed in a row” parameter to a higher value by setting the early argument in the “Optional arguments” to, for example, 15.
The final model will be written to disk as specified in saved_weights_name in your configuration file.
Click to display ⇲
Click to hide ⇱
Navigate to the folder with config_cryolo.json file, train_image folder, etc.
Train your network with 5 warmup epochs in GPU 0:
cryolo_train.py -c config_cryolo.json -w 5 -g 0
The final model file will be written to disk.
5. Picking
Select the action prediction and fill all arguments in the “Required arguments” tab:

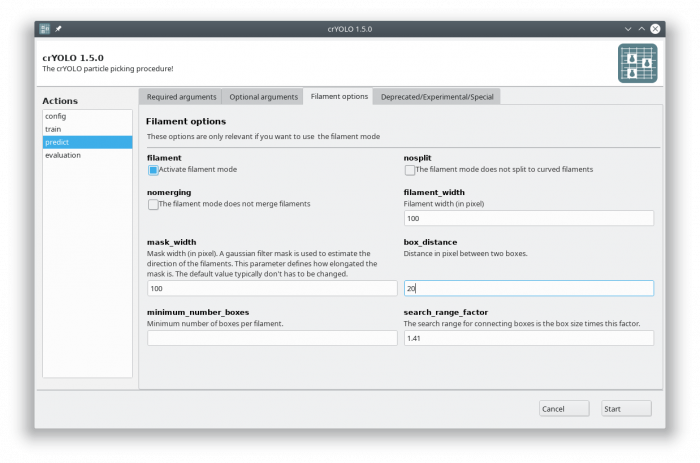
Now select the “Filament options” tab and check “Activate filament mode”, specifiy the filament width (e.g. 100) and define the box distance (e.g. 20 for 90% overlap when using a box size if 200):
The directory output_boxes will be created and all results are saved there. The format is the eman2 helix format with particle coordinates.
You can find a detailed description how to import crYOLO filament coordinates into Relion here.
Alternative: Run prediction in commmand line
Alternative: Run prediction in commmand line
Let's assume you want to pick a filament with a width of 100 pixels (-fw 100). The box size is 200×200 and you want a 90% overlap (-bd 20). Moreover, you wish that each filament has at least 6 boxes (-mn 6). The micrographs are in the full_data directory. Than the picking command would be:
cryolo_predict.py -c cryolo_config.json -w cryolo_model.h5 -i full_data --filament -fw 100 -bd 20 -o boxes/ -g 0 -mn 6
6. Visualize the results
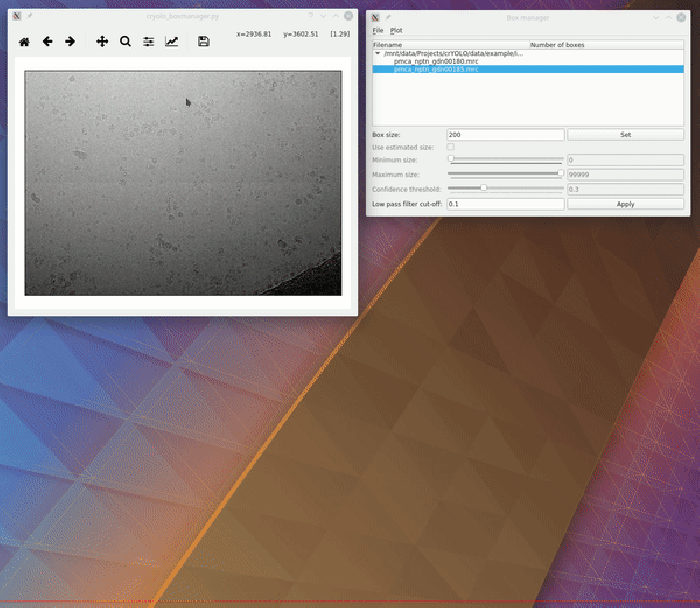
To visualize your results you can use the boxmanager:
As image_dir you select the full_data directory. As box_dir you select the CBOX folder (or EMAN_HELIX_SEGMENTED in case of filaments).
CBOX files contain besides the particle coordinates more information like the confidence and the estimated size of each particle. When importing .cbox files into the box manager, it enables more filtering options in the GUI. You can plot size- and confidence distributions. Moreover, you can change the confidence threshold, minimum and maximum size and see the results in a live preview. If you are done with the filtering, you can then write the new box selection into new box files. The video below shows an example.