Sidebar
Table of Contents
Stripper
STRIPPER: faST Robust fIlament Picking ProcEduRe
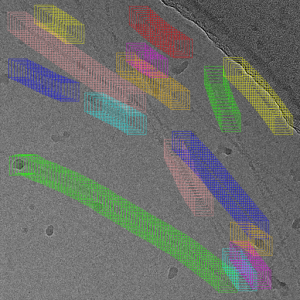
Helps you to strip your filaments out of the micrograph automatically ![]()
Overview
Installation
Activate the “biomedgroup” update site and add a new update site:
http://sites.imagej.net/Mpi-do-stripper/
Get your data into ImageJ
Load the raw data
- File → Import → Image Sequence
- Select the folder of your images
- Type the file ending into “File name contains:” (e.g .mrc, .tif)
- Check “Use virtual stack”
- Press OK Now you have read in your data as an image stack.
Reduce the size
For the sake of memory consumption and speed your should reduce the size / image depth of your image data.
- Image → Adjust → Size
- Set the width to 1024. The height is calculated automatically.
- Press OK.
- Image → Type → 8 Bit Wait until the processing is finished.
Save the file for later use
File → Save As → Tiff…
Parameters
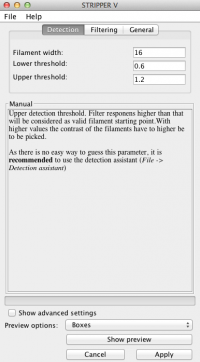
Detection
As there is no easy way to guess the lower and upper threshold, it is recommended to use the detection assistant (File → Detection assistant).
Filament width:
The width of your filaments in pixel.
Mask width (Advanced option):
As the response along the filament randomly fluctuates and sometimes disappear, it is important to average along the filament.
Lower threshold:
Filter responses lower than that value will be considered as background and filament tracing stops here.Higher values lead to more segmented lines, as low contrast regions of a filament will be considered as background the line is splitted at this position.
Upper threshold:
Filter responens higher than that will be considered as valid filament starting point.With higher values the contrast of the filaments have to higher be to be picked.
Equalize:
After enhancement of the filaments, all images are adjusted that filamants have a more similar absolute response. Especially when the image contains high contrast contaminations, this option has a positive effect. By defaut this option is activated.
Filtering
Min number of boxes:
The minimum number of boxes that have to be placed per line.
Response filter
Sigma min. response:
The mean response M and the standard deviation S are estimated over all lines per image. If a line has a signficant number of line points below the threshold T = M-F*S it is considered as false-positiv (background) and is removed, whereas F is value to specifiy.By default, this filter is deactivated (set to 0). A resonable value lies typically between 2 - 3.
Sigma max. response:
The mean response M and the standard deviation S are estimated over all lines per image. If a line has a signficant number of line points above the threshold T = M+F*S it is considered as false-positiv (ice,overlapping filament) and is removed, whereas F is value to specifiy.By default, this filter is deactivated (set to 0). A resonable value lies typically between 2 - 3.
Sensitivity:
Threshold value for minimum straightness. Line segments (see window size) with a straightness below that threshold will be removed and the line is splitted. Higher values means that the filament have to be more straight, where 1 means a perfect straight line.
Straightness filter
Min. straightness:
Threshold value for minimum straightness. Line segments (see window size) with a straightness below that threshold will be removed and the line is splitted. Higher values means that the filament have to be more straight, where 1 means a perfect straight line.
Window size:
The number of line points which are used to estimate the straightness. Larger values lead to more robustness against noisebut decrease to ability to detect small curves.
Allowed box overlap:
Relative amount to what extend two boxes auf different filaments are allowed to overlap. The default value 0.5 which is the maximum value that ensures that a box contains only one filament